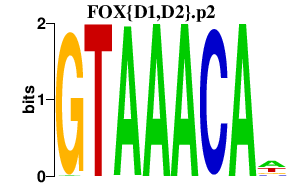
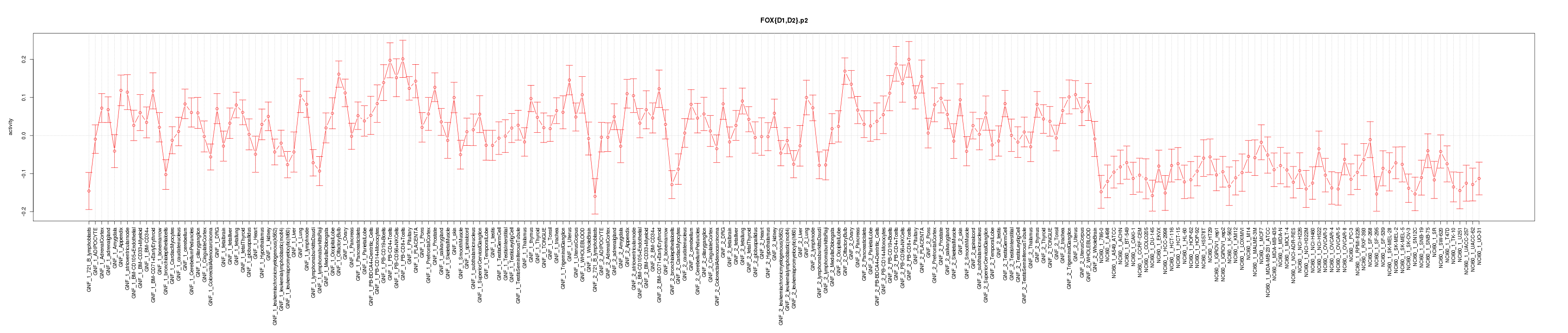
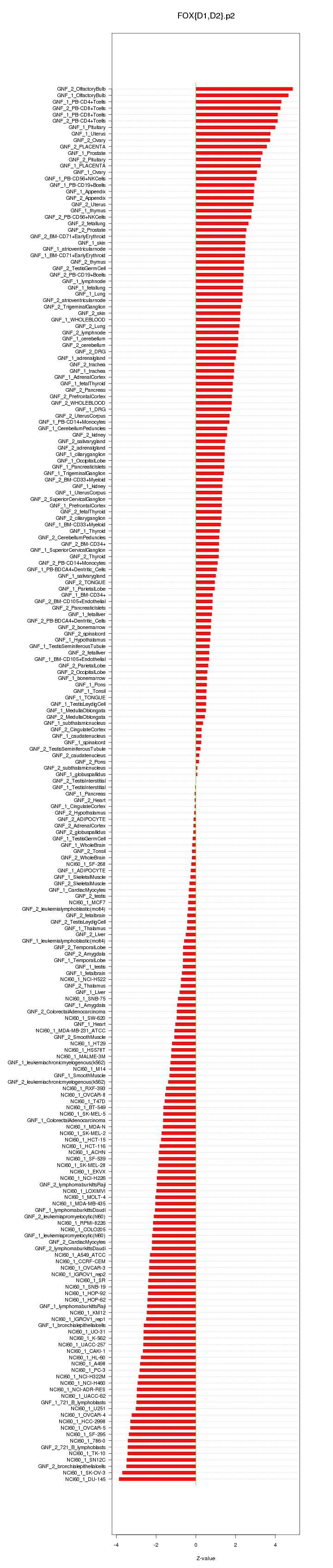
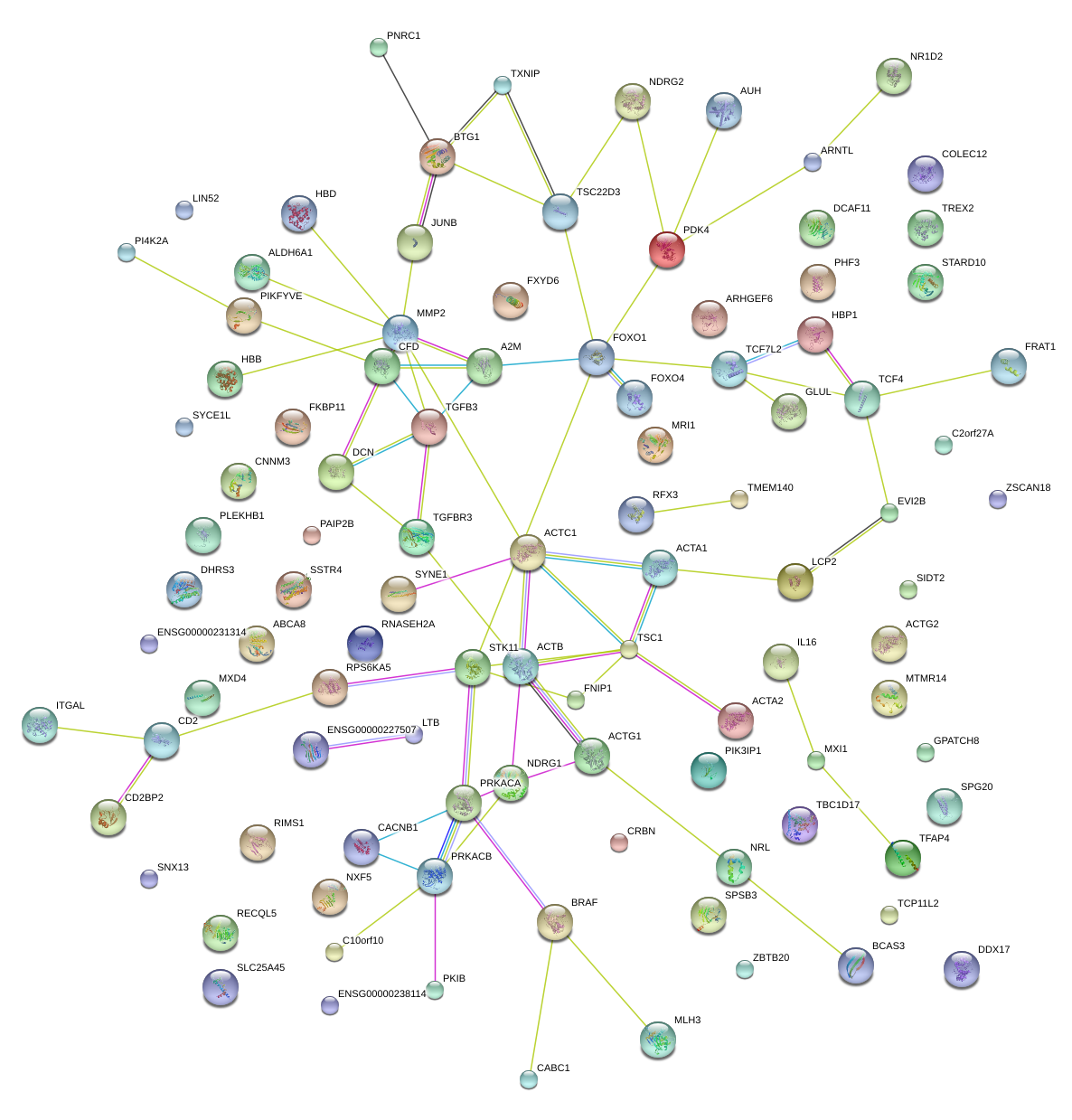
|
chr1_+_145438437
|
44.627
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr5_-_42811959
|
43.006
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr12_-_9268440
|
40.125
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr12_-_92539614
|
37.941
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr6_+_89791510
|
37.935
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr12_-_9268513
|
37.260
|
|
A2M
|
alpha-2-macroglobulin
|
|
chrX_-_106960028
|
35.464
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr12_-_9268522
|
34.934
|
NM_000014
|
A2M
|
alpha-2-macroglobulin
|
|
chrX_-_106960268
|
33.880
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr22_-_31686707
|
33.479
|
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1
|
|
chr12_-_9268475
|
28.611
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr11_-_117747331
|
28.539
|
|
FXYD6
|
FXYD domain containing ion transport regulator 6
|
|
chr17_-_29641068
|
27.366
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr10_-_45474210
|
27.357
|
NM_007021
|
C10orf10
|
chromosome 10 open reading frame 10
|
|
chr13_-_41240607
|
27.268
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr19_+_12902295
|
25.666
|
NM_002229
|
JUNB
|
jun B proto-oncogene
|
|
chr14_+_74551655
|
24.615
|
NM_001024674
|
LIN52
|
lin-52 homolog (C. elegans)
|
|
chr1_+_84630374
|
23.750
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr1_+_117297056
|
23.238
|
NM_001767
|
CD2
|
CD2 molecule
|
|
chr5_-_131132642
|
22.992
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr10_-_90750955
|
22.584
|
NM_001141945
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr14_-_74551159
|
21.839
|
NM_005589
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr11_+_73359734
|
21.481
|
NM_001130036
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr14_-_21493858
|
21.117
|
|
NDRG2
|
NDRG family member 2
|
|
chr5_-_169724614
|
20.937
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chrX_-_101112548
|
20.641
|
NM_032946
|
NXF5
|
nuclear RNA export factor 5
|
|
chr3_+_23987514
|
20.589
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr14_-_74551073
|
20.464
|
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr14_-_21493910
|
20.445
|
NM_016250
NM_201535
NM_201536
NM_201537
|
NDRG2
|
NDRG family member 2
|
|
chr4_-_2263706
|
19.704
|
NM_006454
|
MXD4
|
MAX dimerization protein 4
|
|
chr6_+_122793061
|
19.678
|
NM_181794
|
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta
|
|
chr22_-_31688329
|
19.404
|
NM_001135911
NM_052880
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1
|
|
chr14_+_24584009
|
18.851
|
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr1_-_92351613
|
18.176
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr11_-_5255712
|
17.279
|
NM_000519
|
HBD
|
hemoglobin, delta
|
|
chr12_-_91576805
|
17.106
|
NM_001920
|
DCN
|
decorin
|
|
chr10_+_99079017
|
17.086
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr17_-_29641094
|
17.018
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr17_-_29641087
|
16.959
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chrX_+_70315637
|
16.958
|
NM_001170931
NM_005938
|
FOXO4
|
forkhead box O4
|
|
chr14_-_24584125
|
16.884
|
|
NRL
|
neural retina leucine zipper
|
|
chr16_+_77233348
|
16.548
|
NM_001129979
|
SYCE1L
|
synaptonemal complex central element protein 1-like
|
|
chr5_+_72251869
|
16.444
|
|
|
|
|
chr16_-_1832576
|
16.442
|
NM_080861
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr7_+_106809405
|
16.034
|
NM_001244262
NM_012257
|
HBP1
|
HMG-box transcription factor 1
|
|
chr5_-_169724782
|
16.005
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr1_+_227127993
|
15.857
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr14_+_24584321
|
15.785
|
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr6_-_31550065
|
15.733
|
NM_002341
NM_009588
|
LTB
|
lymphotoxin beta (TNF superfamily, member 3)
|
|
chr7_+_134832765
|
15.607
|
NM_018295
|
TMEM140
|
transmembrane protein 140
|
|
chr1_+_227127959
|
15.514
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr17_+_58755158
|
15.150
|
NM_001099432
NM_017679
|
BCAS3
|
breast carcinoma amplified sequence 3
|
|
chr17_-_66951381
|
14.971
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr12_+_106696508
|
14.956
|
NM_152772
|
TCP11L2
|
t-complex 11 (mouse)-like 2
|
|
chr16_+_30484049
|
14.934
|
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide)
|
|
chr7_-_140624280
|
14.903
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr18_-_53257033
|
14.848
|
NM_001243227
NM_001243228
|
TCF4
|
transcription factor 4
|
|
chr18_-_53303129
|
14.801
|
NM_001243226
|
TCF4
|
transcription factor 4
|
|
chr19_-_58609606
|
14.786
|
NM_001145543
NM_001145544
NM_023926
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr22_-_38902344
|
14.681
|
NM_001098504
NM_006386
|
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17
|
|
chr19_-_58609574
|
14.397
|
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr5_-_169724734
|
14.298
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr2_+_74120132
|
14.263
|
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr11_+_13299260
|
14.253
|
NM_001030272
NM_001030273
NM_001178
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr10_+_99400469
|
14.243
|
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha
|
|
chr11_-_65150107
|
14.242
|
NM_001077241
NM_182556
|
SLC25A45
|
solute carrier family 25, member 45
|
|
chr11_+_117049838
|
14.213
|
NM_001040455
|
SIDT2
|
SID1 transmembrane family, member 2
|
|
chr14_-_76447445
|
14.089
|
|
TGFB3
|
transforming growth factor, beta 3
|
|
chr14_-_91526912
|
14.046
|
NM_004755
NM_182398
|
RPS6KA5
|
ribosomal protein S6 kinase, 90kDa, polypeptide 5
|
|
chr1_+_227127999
|
14.039
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr9_-_3525973
|
14.029
|
NM_002919
NM_134428
|
RFX3
|
regulatory factor X, 3 (influences HLA class II expression)
|
|
chr2_+_97481977
|
13.991
|
NM_017623
NM_199078
|
CNNM3
|
cyclin M3
|
|
chr5_-_169724817
|
13.930
|
NM_005565
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr14_-_76448091
|
13.877
|
NM_003239
|
TGFB3
|
transforming growth factor, beta 3
|
|
chr7_-_17980102
|
13.691
|
NM_015132
|
SNX13
|
sorting nexin 13
|
|
chr16_+_30483959
|
13.640
|
NM_001114380
NM_002209
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide)
|
|
chr8_-_134309448
|
13.424
|
NM_001135242
NM_006096
|
NDRG1
|
N-myc downstream regulated 1
|
|
chr1_-_12677347
|
13.371
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr15_+_81589240
|
13.359
|
NM_004513
|
IL16
|
interleukin 16
|
|
chr16_-_1832571
|
13.359
|
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr12_-_91576579
|
13.322
|
|
DCN
|
decorin
|
|
chr19_+_50380681
|
13.224
|
NM_001168222
NM_024682
|
TBC1D17
|
TBC1 domain family, member 17
|
|
chr2_+_74120092
|
13.195
|
NM_001199893
NM_001615
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr19_+_859658
|
13.160
|
NM_001928
|
CFD
|
complement factor D (adipsin)
|
|
chr11_+_13299391
|
13.110
|
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr10_+_111985628
|
12.923
|
NM_005962
|
MXI1
|
MAX interactor 1
|
|
chr14_+_24583979
|
12.862
|
NM_025230
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr1_+_227127937
|
12.847
|
NM_020247
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr14_-_75518132
|
12.788
|
NM_001040108
NM_014381
|
MLH3
|
mutL homolog 3 (E. coli)
|
|
chr9_-_135819995
|
12.762
|
NM_000368
NM_001162426
NM_001162427
|
TSC1
|
tuberous sclerosis 1
|
|
chr11_+_13299376
|
12.678
|
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr11_+_13299372
|
12.668
|
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chrX_-_135863502
|
12.662
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr14_+_24583905
|
12.607
|
NM_001163484
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr16_+_55512801
|
12.587
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr16_-_4322695
|
12.540
|
|
TFAP4
|
transcription factor AP-4 (activating enhancer binding protein 4)
|
|
chr16_-_86542463
|
12.477
|
|
LOC400550
|
uncharacterized LOC400550
|
|
chr18_-_500583
|
12.368
|
NM_130386
|
COLEC12
|
collectin sub-family member 12
|
|
chr9_-_94124149
|
12.355
|
|
AUH
|
AU RNA binding protein/enoyl-CoA hydratase
|
|
chr1_-_92351476
|
12.311
|
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr20_+_23015949
|
12.283
|
NM_001052
|
SSTR4
|
somatostatin receptor 4
|
|
chr6_+_64345726
|
12.218
|
|
PHF3
|
PHD finger protein 3
|
|
chr19_+_859673
|
12.196
|
|
CFD
|
complement factor D (adipsin)
|
|
chr15_-_44828864
|
12.176
|
|
|
|
|
chr2_-_71454200
|
12.165
|
NM_020459
|
PAIP2B
|
poly(A) binding protein interacting protein 2B
|
|
chr11_+_13299349
|
12.047
|
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr7_-_95225802
|
12.004
|
NM_002612
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4
|
|
chr3_-_114343052
|
11.934
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr6_+_72922513
|
11.907
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr6_+_64345709
|
11.858
|
|
PHF3
|
PHD finger protein 3
|
|
chr10_+_111969988
|
11.844
|
NM_001008541
|
MXI1
|
MAX interactor 1
|
|
chr19_+_1205773
|
11.711
|
NM_000455
|
STK11
|
serine/threonine kinase 11
|
|
chr1_-_182357871
|
11.709
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr16_-_1832304
|
11.579
|
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr2_+_132480063
|
11.529
|
NM_013310
|
C2orf27A
|
chromosome 2 open reading frame 27A
|
|
chr13_-_36920396
|
11.523
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr3_-_3221390
|
11.428
|
NM_001173482
NM_016302
|
CRBN
|
cereblon
|
|
chr6_+_64345672
|
11.409
|
|
PHF3
|
PHD finger protein 3
|
|
chr19_+_13875345
|
11.383
|
|
MRI1
|
methylthioribose-1-phosphate isomerase homolog (S. cerevisiae)
|
|
chr11_-_72504643
|
11.368
|
NM_006645
|
STARD10
|
StAR-related lipid transfer (START) domain containing 10
|
|
chr3_+_9691184
|
11.318
|
|
MTMR14
|
myotubularin related protein 14
|
|
chr3_-_3221361
|
11.311
|
|
CRBN
|
cereblon
|
|
chr3_-_3221385
|
11.278
|
|
CRBN
|
cereblon
|
|
chr17_-_42580745
|
11.099
|
|
GPATCH8
|
G patch domain containing 8
|
|
chr6_-_152623213
|
11.003
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr1_-_154840460
|
10.981
|
|
|
|
|
chr17_-_40897025
|
10.973
|
NM_001991
|
EZH1
|
enhancer of zeste homolog 1 (Drosophila)
|
|
chr22_-_38902315
|
10.945
|
|
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17
|
|
chr1_-_45956795
|
10.932
|
NM_007170
|
TESK2
|
testis-specific kinase 2
|
|
chr22_-_38902281
|
10.920
|
|
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17
|
|
chr1_-_115053677
|
10.778
|
|
TRIM33
|
tripartite motif containing 33
|
|
chr14_-_21493122
|
10.775
|
NM_201538
NM_201539
NM_201540
NM_201541
|
NDRG2
|
NDRG family member 2
|
|
chr5_+_176853673
|
10.747
|
NM_001004105
NM_001004106
NM_002082
|
GRK6
|
G protein-coupled receptor kinase 6
|
|
chr5_-_146435589
|
10.695
|
NM_181674
NM_181676
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr3_+_46411632
|
10.618
|
NM_000579
NM_001100168
|
CCR5
|
chemokine (C-C motif) receptor 5
|
|
chr3_+_147127151
|
10.471
|
NM_003412
|
ZIC1
|
Zic family member 1
|
|
chr5_-_64920100
|
10.464
|
|
TRIM23
|
tripartite motif containing 23
|
|
chr3_-_48956817
|
10.397
|
NM_001123040
|
C3orf71
|
chromosome 3 open reading frame 71
|
|
chr1_+_209878135
|
10.363
|
NM_005525
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr9_+_88897112
|
10.305
|
|
|
|
|
chr10_+_51549552
|
10.264
|
NM_002443
NM_138634
|
MSMB
|
microseminoprotein, beta-
|
|
chr20_+_48429375
|
10.213
|
|
SLC9A8
|
solute carrier family 9 (sodium/hydrogen exchanger), member 8
|
|
chr3_+_9691116
|
9.985
|
NM_001077525
NM_001077526
NM_022485
|
MTMR14
|
myotubularin related protein 14
|
|
chr12_-_49504337
|
9.980
|
|
LMBR1L
|
limb region 1 homolog (mouse)-like
|
|
chr22_-_38902300
|
9.963
|
|
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17
|
|
chr11_+_10476665
|
9.955
|
NM_001025389
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr6_+_134758840
|
9.812
|
|
LOC154092
|
uncharacterized LOC154092
|
|
chr6_-_33168369
|
9.805
|
NM_021976
|
RXRB
|
retinoid X receptor, beta
|
|
chr13_+_79980420
|
9.757
|
|
RBM26-AS1
|
RBM26 antisense RNA 1 (non-protein coding)
|
|
chr15_-_40600118
|
9.755
|
NM_004573
|
PLCB2
|
phospholipase C, beta 2
|
|
chr11_+_63606535
|
9.730
|
|
MARK2
|
MAP/microtubule affinity-regulating kinase 2
|
|
chr17_+_65373923
|
9.695
|
NM_012417
NM_181671
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1
|
|
chr17_+_77751929
|
9.593
|
NM_005189
NM_032647
|
CBX2
|
chromobox homolog 2
|
|
chr14_+_56585147
|
9.550
|
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr5_-_64920149
|
9.536
|
NM_001656
NM_033227
NM_033228
|
TRIM23
|
tripartite motif containing 23
|
|
chr1_-_182357757
|
9.430
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr7_-_115670774
|
9.371
|
|
TFEC
|
transcription factor EC
|
|
chr11_+_13299329
|
9.351
|
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr7_-_76829109
|
9.269
|
NM_006682
|
FGL2
|
fibrinogen-like 2
|
|
chr7_-_37393271
|
9.241
|
NM_001206482
|
ELMO1
|
engulfment and cell motility 1
|
|
chr3_+_9691198
|
9.240
|
|
MTMR14
|
myotubularin related protein 14
|
|
chr12_+_96588254
|
9.226
|
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2)
|
|
chr15_+_99191671
|
9.177
|
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr12_+_12764755
|
9.148
|
NM_001310
|
CREBL2
|
cAMP responsive element binding protein-like 2
|
|
chr16_-_68033355
|
9.109
|
NM_022355
|
DPEP2
|
dipeptidase 2
|
|
chr5_+_169064238
|
9.105
|
NM_004946
|
DOCK2
|
dedicator of cytokinesis 2
|
|
chr17_-_80656461
|
9.025
|
NM_006822
|
RAB40B
|
RAB40B, member RAS oncogene family
|
|
chr2_+_12856813
|
8.977
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr19_-_50316360
|
8.926
|
NM_001171937
NM_025129
|
FUZ
|
fuzzy homolog (Drosophila)
|
|
chr11_+_63606346
|
8.756
|
NM_001039469
NM_001163296
NM_001163297
NM_004954
|
MARK2
|
MAP/microtubule affinity-regulating kinase 2
|
|
chr8_+_22133079
|
8.687
|
NM_001135721
|
PIWIL2
|
piwi-like 2 (Drosophila)
|
|
chr7_+_65419441
|
8.673
|
|
VKORC1L1
|
vitamin K epoxide reductase complex, subunit 1-like 1
|
|
chr6_+_111303240
|
8.663
|
NM_032194
|
RPF2
|
ribosome production factor 2 homolog (S. cerevisiae)
|
|
chr1_+_150122415
|
8.592
|
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr2_-_198175324
|
8.571
|
|
ANKRD44
|
ankyrin repeat domain 44
|
|
chr7_-_17979998
|
8.495
|
|
SNX13
|
sorting nexin 13
|
|
chr8_+_22132809
|
8.465
|
NM_018068
|
PIWIL2
|
piwi-like 2 (Drosophila)
|
|
chrX_-_15333726
|
8.456
|
NM_001201583
NM_080873
|
ASB11
|
ankyrin repeat and SOCS box containing 11
|
|
chr14_-_23288912
|
8.440
|
NM_001126106
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chr1_-_20306908
|
8.422
|
NM_000300
NM_001161727
NM_001161728
|
PLA2G2A
|
phospholipase A2, group IIA (platelets, synovial fluid)
|
|
chr3_-_12941547
|
8.421
|
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr15_-_60884615
|
8.405
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr11_+_63606598
|
8.361
|
|
MARK2
|
MAP/microtubule affinity-regulating kinase 2
|
|
chr8_-_127570465
|
8.219
|
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr1_+_150122095
|
8.204
|
NM_016274
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr15_+_25230199
|
8.189
|
|
PAR5
|
Prader-Willi/Angelman syndrome-5
|
|
chr16_-_4323000
|
8.180
|
NM_003223
|
TFAP4
|
transcription factor AP-4 (activating enhancer binding protein 4)
|
|
chr1_+_150122246
|
8.116
|
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr5_-_142065991
|
8.091
|
NM_033136
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr6_+_139094656
|
8.008
|
NM_015439
|
CCDC28A
|
coiled-coil domain containing 28A
|
|
chr11_-_66335958
|
7.975
|
NM_003793
|
CTSF
|
cathepsin F
|
|
chr11_-_114466397
|
7.945
|
|
FAM55D
|
family with sequence similarity 55, member D
|
|
chr6_-_87804773
|
7.889
|
NM_000735
NM_001252383
|
CGA
|
glycoprotein hormones, alpha polypeptide
|
|
chr19_-_38720312
|
7.766
|
NM_001135156
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr10_-_90712510
|
7.761
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr14_+_56584854
|
7.676
|
NM_021255
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr12_-_48213567
|
7.657
|
NM_001098416
NM_015401
|
HDAC7
|
histone deacetylase 7
|
|
chr16_-_57318574
|
7.623
|
NM_015993
|
PLLP
|
plasmolipin
|
|
chr17_+_38219040
|
7.615
|
NM_001190918
NM_003250
NM_199334
|
THRA
|
thyroid hormone receptor, alpha
|
|
chr19_+_57752051
|
7.599
|
NM_001023563
NM_001145078
|
ZNF805
|
zinc finger protein 805
|